Melanocytes develop from multipotent neural crest cells which via multiple state transitions eventually gives rise to a mature melanocyte that imparts its function of photoprotection to the skin and the hair follicle. There are broadly two kinds of transitions happening here: developmental context and functional (homeostatic) context. My work involves understanding both these transitions in detail.
Unravelling Melanocyte Heterogeneity using Single Cell Methodologies Melanocytes respond to UV radiation by producing melanin, initially promoting cell survival through increased proliferation and later transferring melanin to neighboring keratinocytes for protection. To understand if the same cells activate these divergent programs of pigmentation and proliferation and if the extent of activation varies within the melanocyte population, we employ single-cell techniques, including sequencing and imaging. These methods allow us to analyze gene expression and visualize cellular processes at the individual cell level. By correlating gene expression patterns with phenotypic changes, we aim to uncover the mechanisms underlying melanocyte's dual response. This research may lead to new insights into skin protection strategies against UV-induced damage and skin cancer. The manuscript is uploaded on bioRxiv and is currently in communication.
Elated to finally share my PhD work 🤩 uncovering the shades of grey in the black & white lives of melanocytes. Huge thanks to everyone involved @PigmentCellBio @rajesh_gokhale @mkjolly15 @IGIBSocial https://t.co/uWmBzTnNxO
— Ayush Aggarwal (@AggarwalAyush_) May 10, 2023
A🧵to explain the findings briefly.
1/n
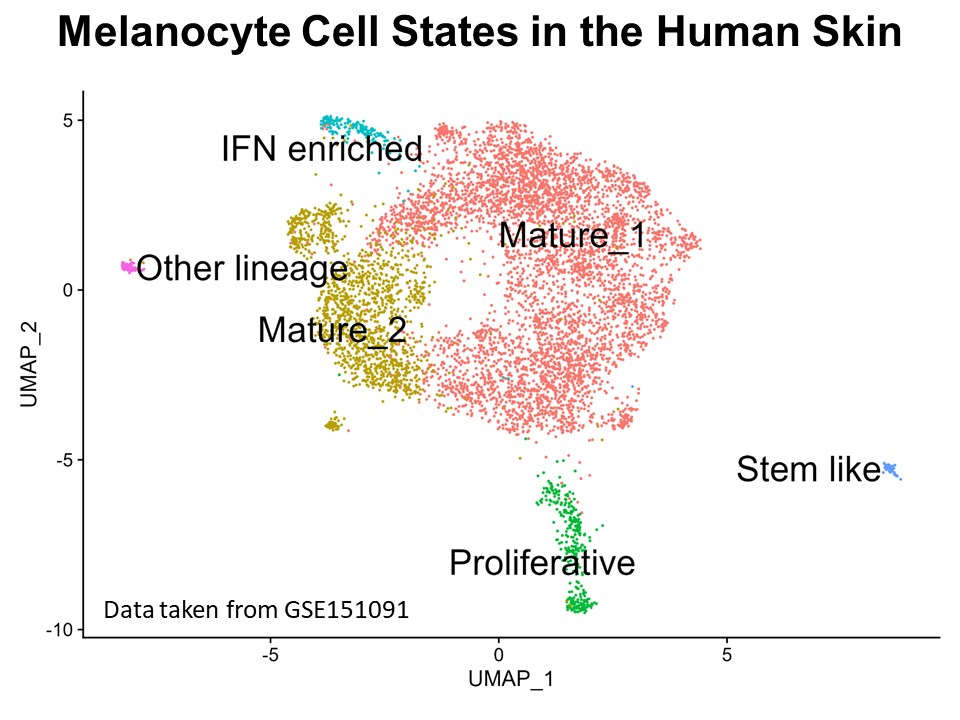
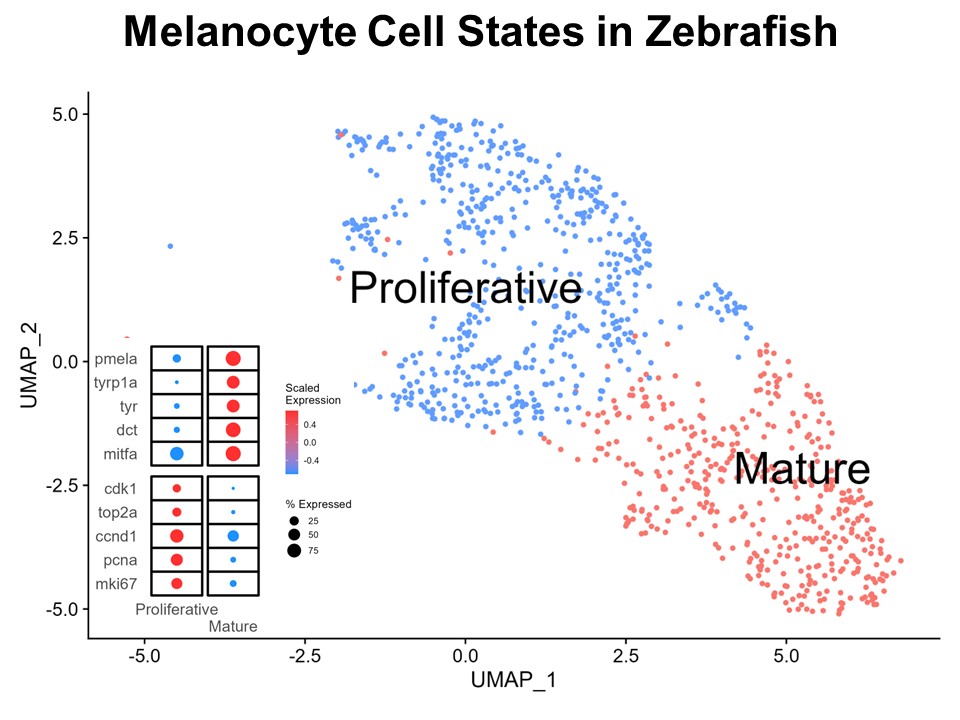
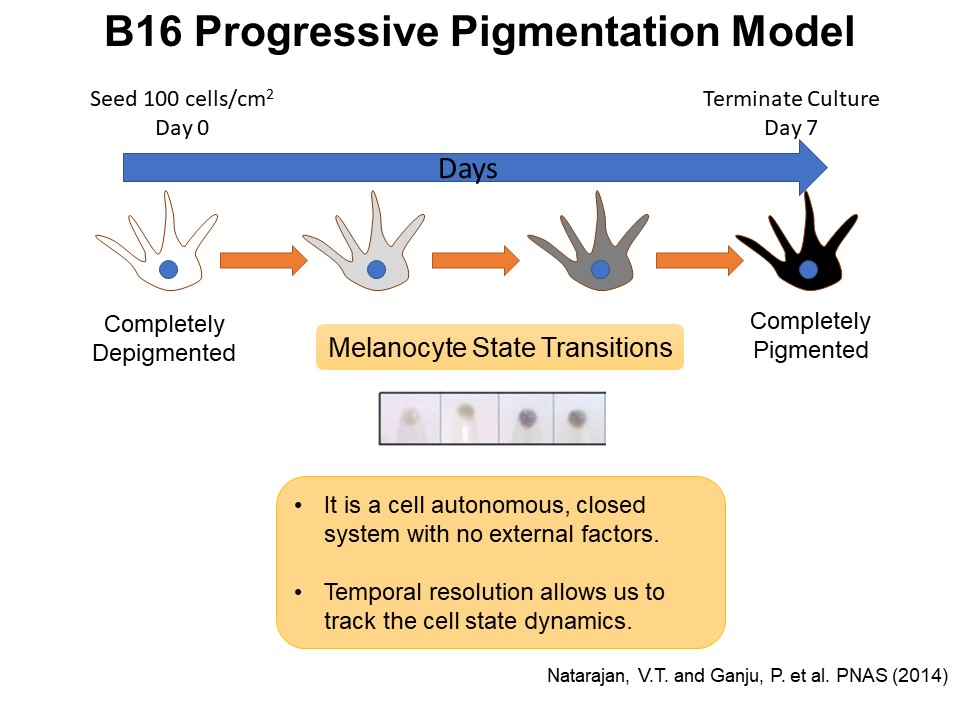
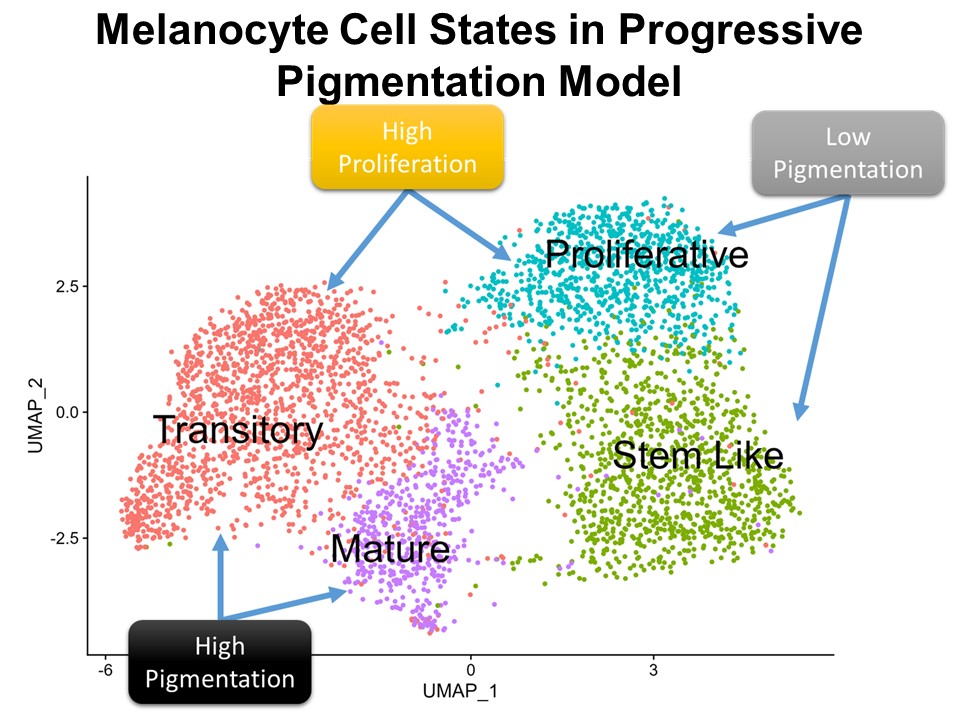

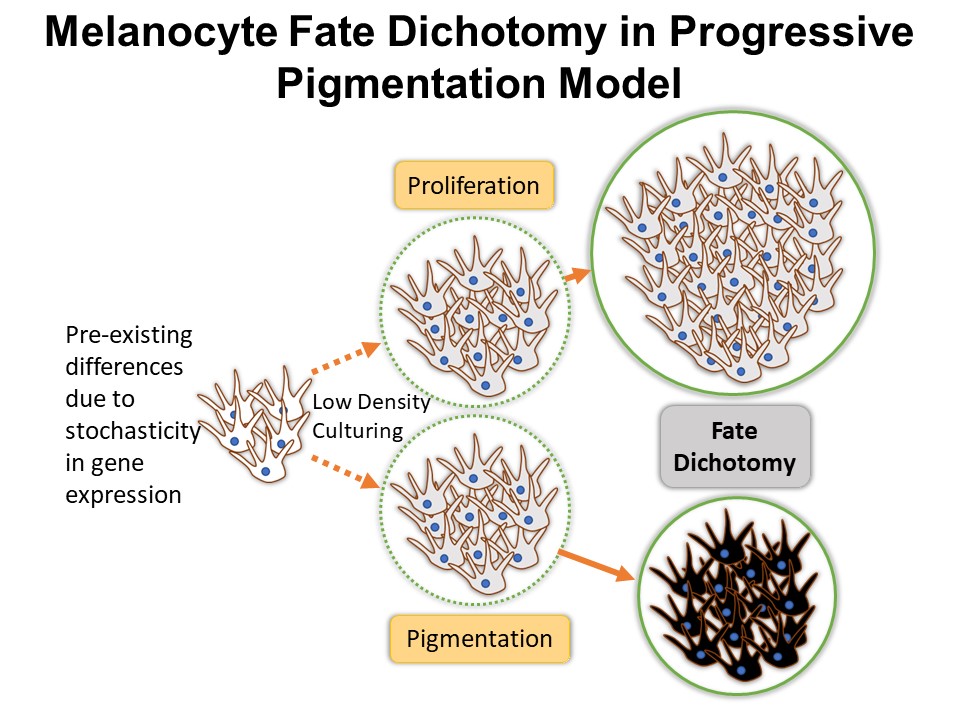
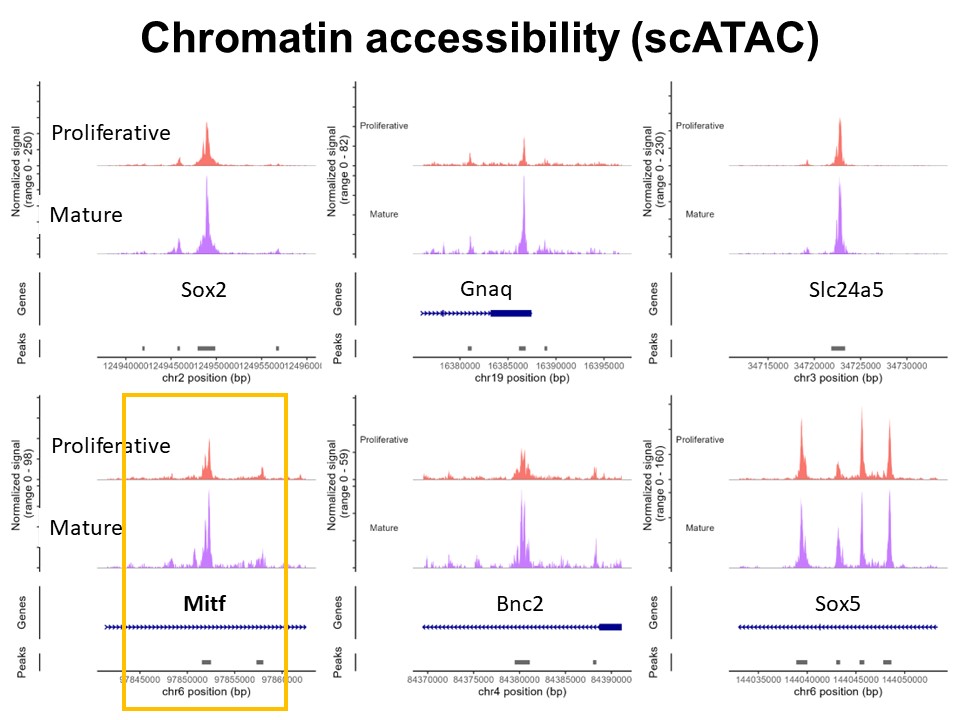
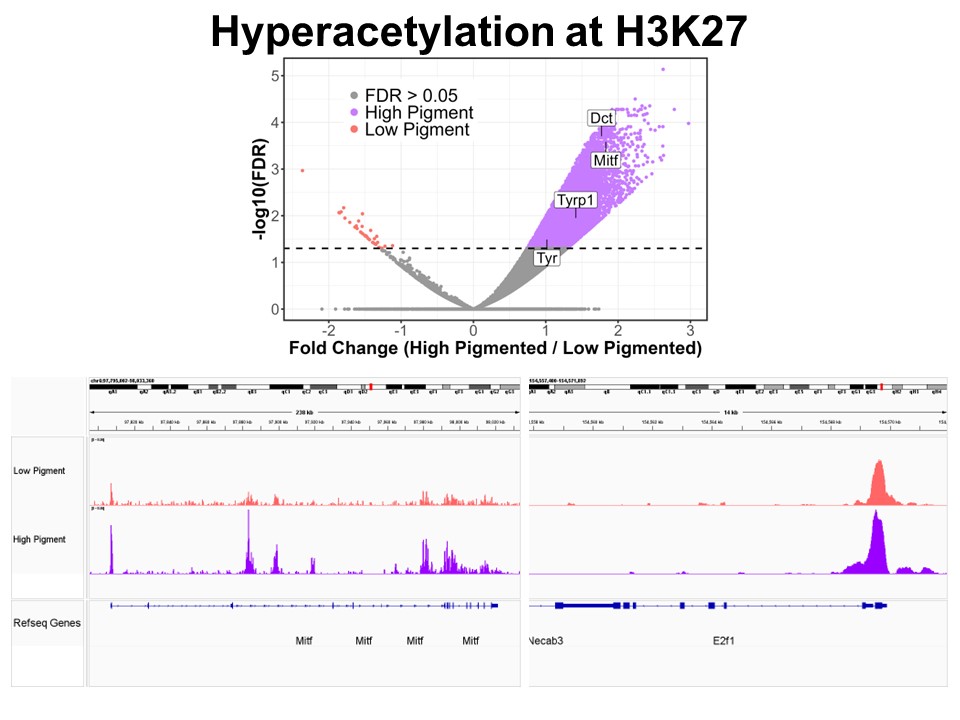
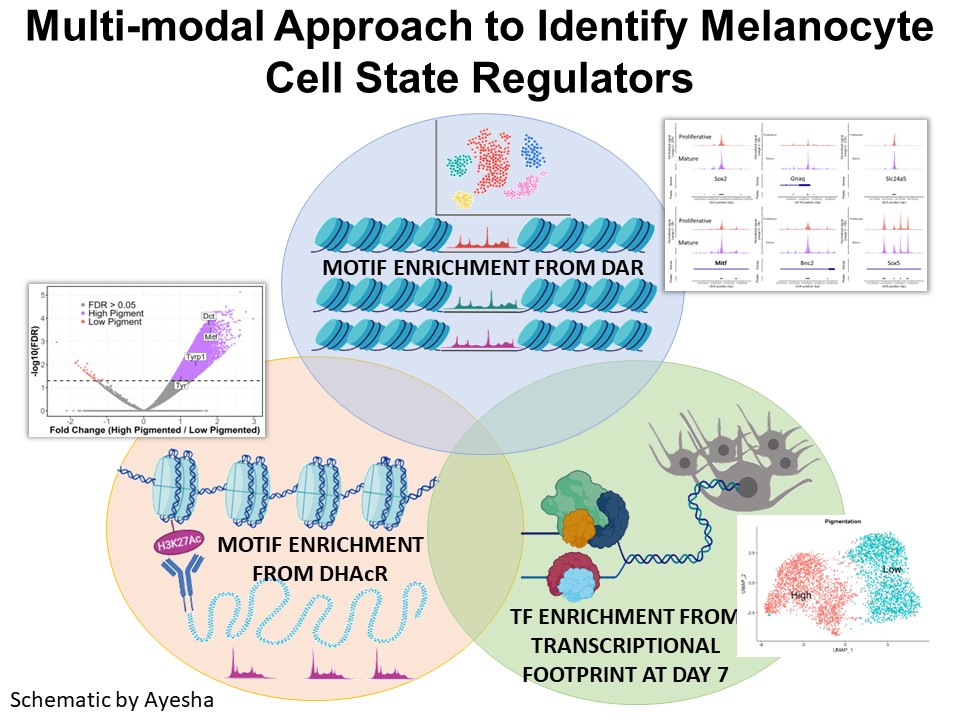
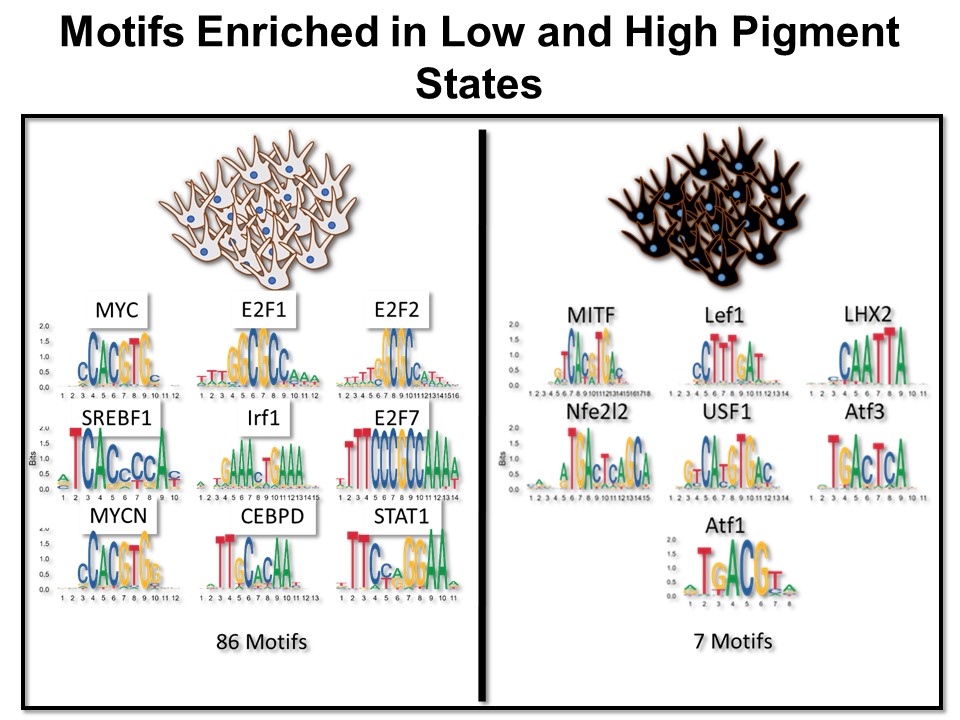
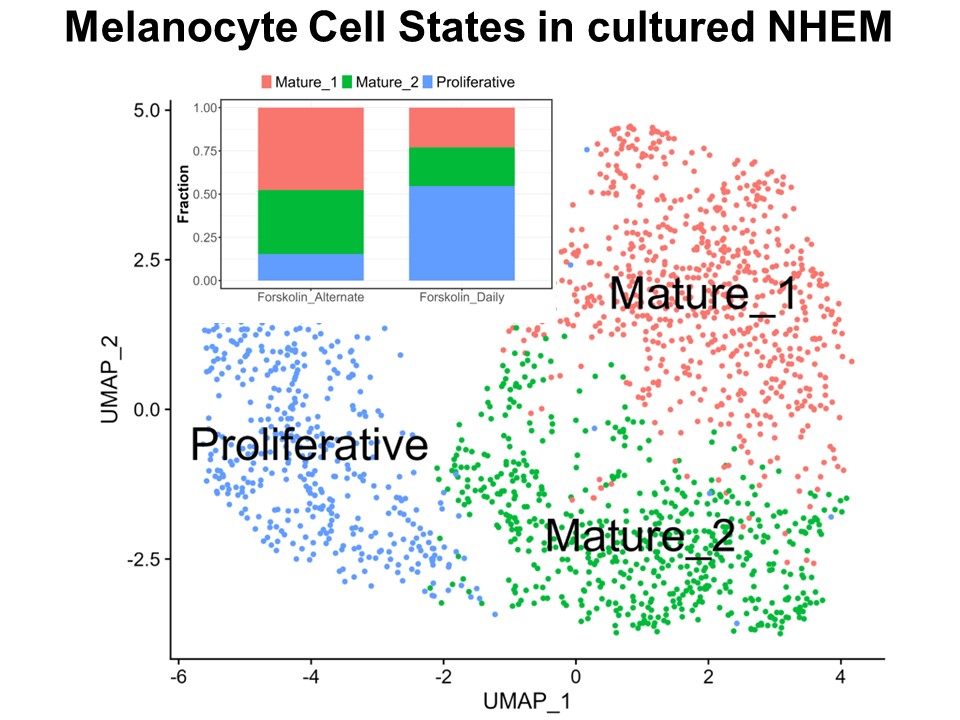

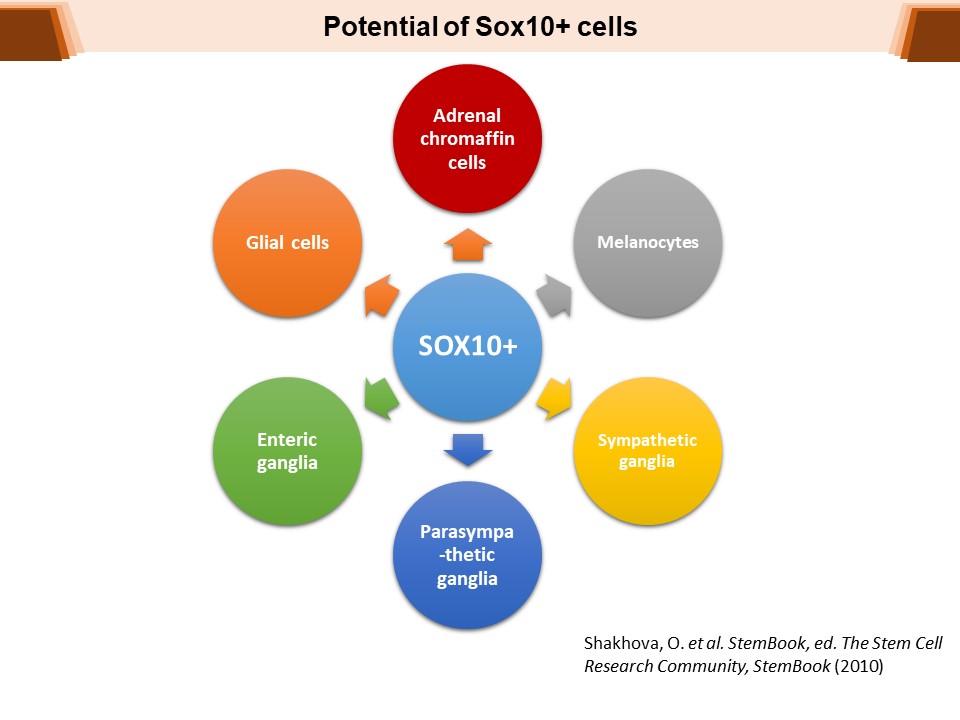
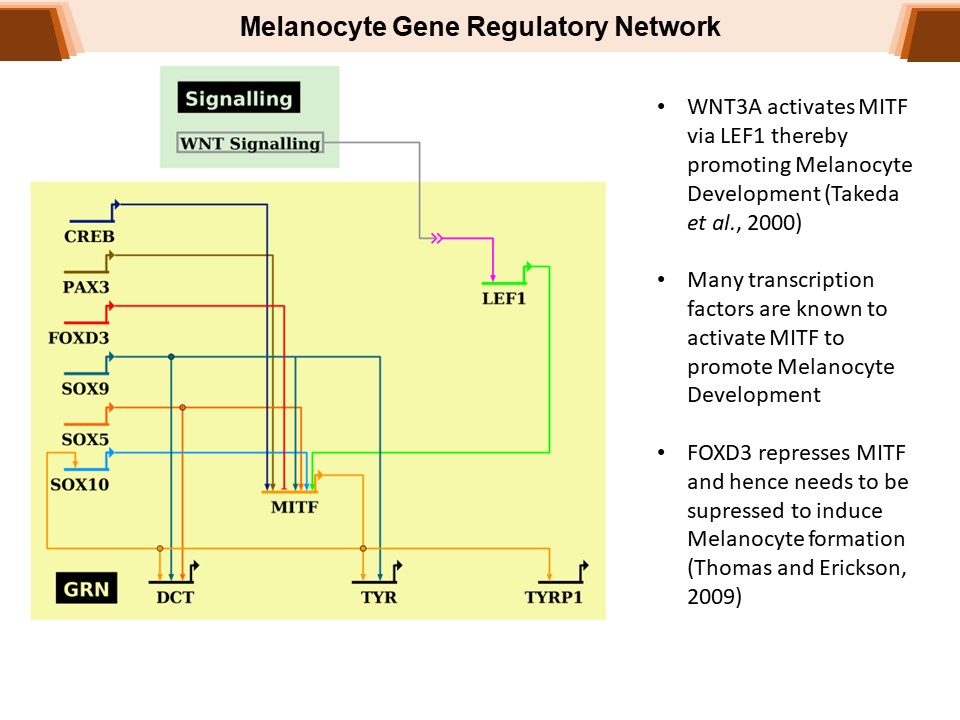
Identification of Novel Regulators of Melanocyte Development using a Data Driven Approach Melanocytes derive from multipotent neural crest cells through a sox10+ progenitor, which possesses the ability to differentiate into various cell types, including melanocytes. Understanding the specific gene network responsible for directing the sox10+ cells towards the melanocyte fate is crucial. Although some genes are already known to bias the multipotent progenitor towards melanocyte lineage, there is still a vast potential for identifying novel regulators and expanding our knowledge of melanocyte development. In my research, I leverage multiple publicly available datasets that capture melanocyte development to identify these regulators and shed light on the intricate process of melanocyte development. The manuscript for this part is currently under preparation.
Introducing MelDat: A Web-Based Application for Exploring Melanocyte Datasets. I have developed an R Shiny-based application that provides a user-friendly platform for effortless exploration of multiple melanocyte-related datasets. Originally conceived as a resource for our laboratory, it initially contained datasets generated within our research group. During our lab meetings and scientific discussions, we often encountered new genes that showed potential relevance to melanocyte biology. We immediately became intrigued to understand the behavior of these genes within our datasets and their potential role in melanocytes. In order to foster and sustain this curiosity, and to eliminate the need for repeated manual exploration of the datasets, I developed this web-based application. Eventually, I expanded the application to include other publicly available datasets as well, and I plan to continue incorporating more as time permits. What's more, MelDat provides the capability to explore your own RNA sequencing or microarray datasets, generating high-quality figures suitable for publication. This ongoing effort aims to provide researchers with a comprehensive tool to investigate and analyze melanocyte datasets conveniently. Now, researchers can easily access and analyze these diverse datasets, empowering them to delve deeper into this fascinating field of study.
